What is stored in MolMeDB?
MolMeDB was developed as centralized storage of drug-membrane and drug-transporter interactions. These data are stored in a uniform format and offered to users for free via the web interface or REST API. The application also serves several tools to improve the data quality, such as an auto-validator trying to match saved molecules with their corresponding records in other databases (Pubchem, PDB, DrugBank, ChEMBL or ChEBI). Each molecule is uniquely determined by MolMeDB identifier and SMILES notation describing its structure. Before the auto-validator run is firstly computed molecular weight and LogP values using RDKit software, and SMILES descriptor is canonized. The molecular structure is subsequently analysed for the presence of functional groups, and closely-related molecules (differing in presence/absence of a functional group) are then grouped for a detailed analysis of the influence of functional groups in a given molecule. These analyses require a sufficient number of molecules and their interactions stored in the database.
Passive interactions
Small molecule interaction with a biological membrane is called passive interaction due to the non-involvement of any transmembrane carrier. The information about passive interactions is gathered in the section “passive interaction” on the compound detail page of the molecule. To thoroughly describe interaction details, we are collecting several properties, which are dependent on the used method (see section methods). A complete list of properties defined in the section “passive interactions” is provided below:
General properties
- T - temperature [°C],
- Q - charge of the molecule,
- LogP - logarithm of a partitioning coefficient octanol-water [molo/molw],
- MW - molecular weight [Da].
Properties related to the permeability methods
- LogPerm - logarithm of a permeability coefficient [cm/s],
- Xmin - drug position with minimum energy in the membrane [nm],
- LogK - logarithm of a partitioning coefficient membrane-water [molm/molw],
- ΔGpen - penetration barrier [kcal/mol], and
- ΔGwat - affinity towards the membrane [kcal/mol].
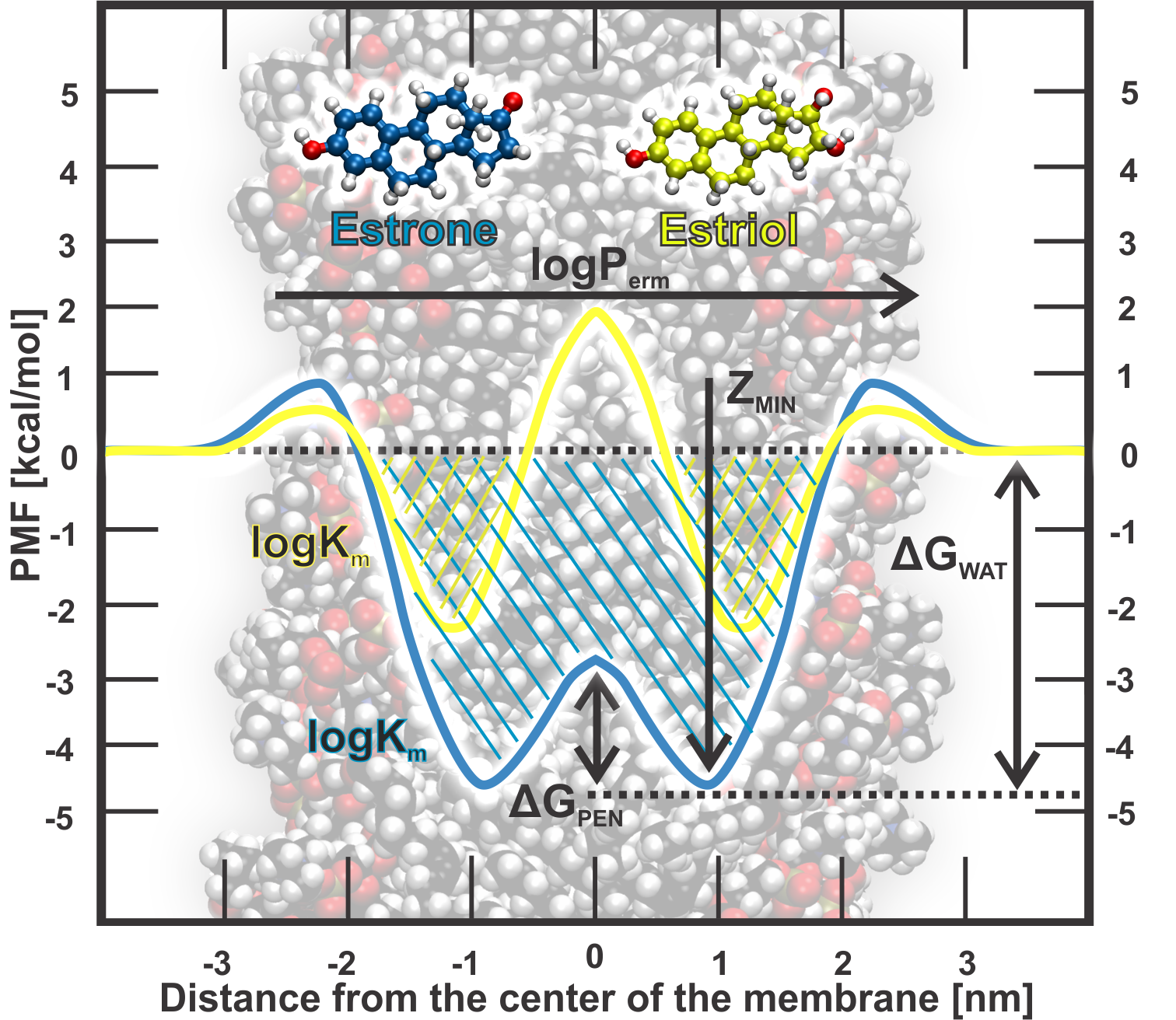
Figure 1. Summary of all properties obtainable from a free energy profile: LogPerm, Xmin (Zmin), ΔGwat, ΔGpen and LogK.
Properties related to spectroscopic methods
- It - Fluorescent lifetime [ns],
- Fluo_wl - Peak fluorescent wavelength [nm],
- Abs_wl - Peak absorption wavelength [nm].
Other properties
- Theta - contact angle in membrane [°],
- Free energy (PMF) profile along the membrane normal - energetical information about permeation process [kcal/mol along nm]
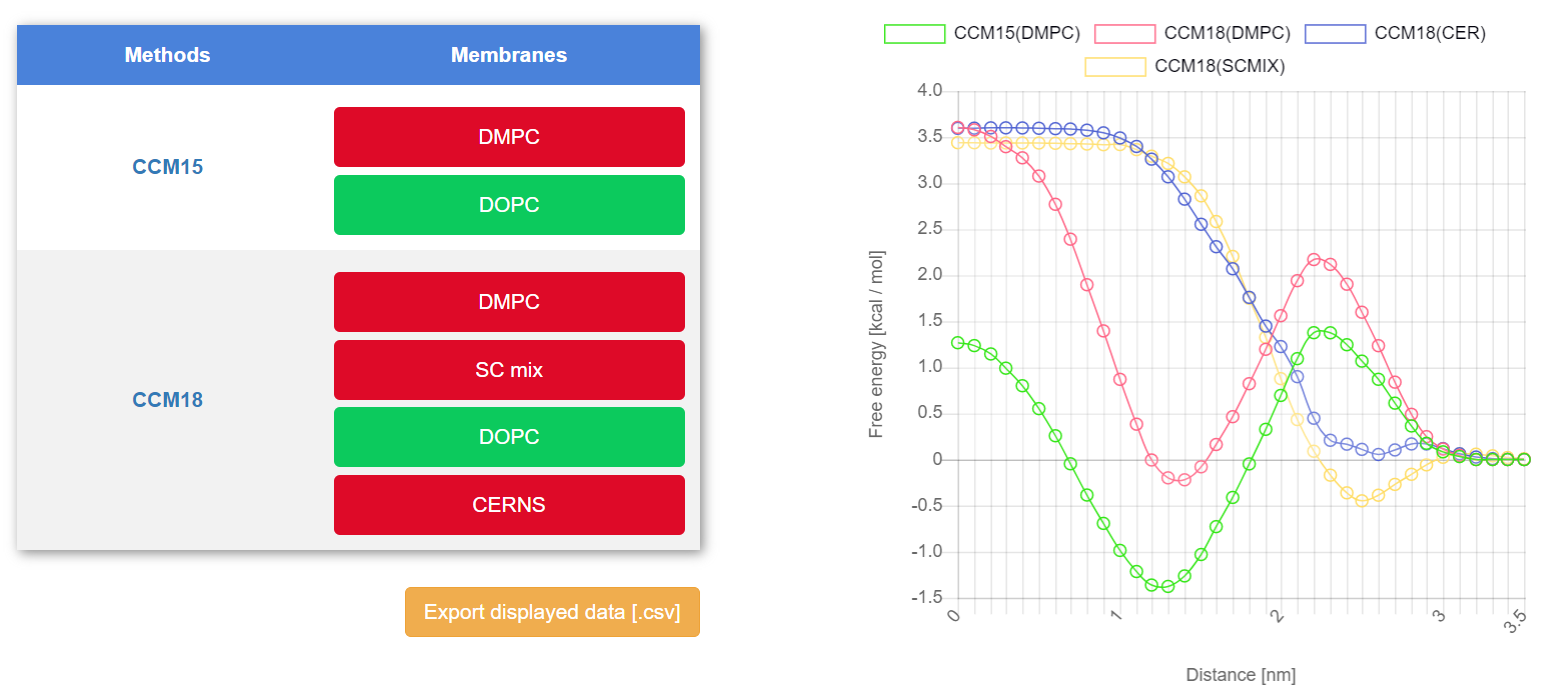
Figure 2. PMF profile of caffeine molecule with interactive options of methods and membranes selection.
Note,
All passive interactions are organised into datasets with a common secondary reference, whereas the primary reference is always directly related to the corresponding interaction. As secondary is always a labelled reference, where our team collected the data, but, on the contrary, a primary reference corresponds to the original data source.
Methods
Currently (01/2023), MolMeDB contains 54 different methods that allow the study of the behaviour of molecules on membranes. Because of better clarity, the classification system for methods was defined, including three main categories of methods – experimental, calculated, and simulated. Each main category is divided into one or more subcategories. Table 1 shows particular categories and subcategories. For an updated list of methods and their categories, please see https://molmedb.upol.cz/browse/methods.
Table 1. Classification of passive interactions methods in MolMeDB.
| Main category | Subcategory |
|---|---|
| Experimental | Partitioning |
| Permeability | |
| Positioning | |
| Calculated | QSAR |
| Quantum mechanics-based | |
| Mechanistic | |
| Simulated | Atomistic MD |
| Hybrid MD | |
| United atoms MD | |
| Coarse-grained MD |
Experimental methods include various types of lab methods, e.g. spectroscopic and microscopic methods or permeability assays. According to measured properties, methods are divided into three subcategories – partitioning, permeability and positioning. Methods classified as partitioning are related to the partitioning coefficient (LogK) of compounds on the membranes. The outputs of experimental “permeability” methods are related to the permeability coefficient (LogPerm). The positioning methods…
Calculated methods are referred to in three subcategories: QSAR, quantum mechanics-based (Q-based) and mechanistic approaches. Subcategory “QSAR” includes data from several QSAR models related to the permeability coefficient prediction (LogPerm). Within MolMeDB is one quantum mechanics-based method – 1COSMOmic, 2COSMOmic/COSMOperm. This method provides several properties about the behaviour of the compound on the membrane (LogPerm, LogK, Xmin, ΔGpen and ΔGwater) and, additionally, part of the results of the COSMOmic or COSMOmic/COSMOperm method is a free energy profile (PMF) of molecules. The PMF is available in the section Free energy graph on a compound page of a molecule (Figure 2). As the mechanistic method is referred a PerMM model and, like the other calculated methods, predicts the permeability coefficient (LogPerm).
The last major category contains simulated methods, including various types of molecular dynamics methods. The subcategories are defined according to granularity levels of membranes as atomistic, hybrid, United-atoms and coarse-grained. This category can provide information on various properties, e.g. LogPerm, Xmin, ΔGpen, ΔGwater, and theta… .
1COSMOmic means COSMOmic software in a version that was published in 2015.
2COSMOmic/COSMOperm has the meaning of COSMOmic software in version 2018. Within the new version, the calculation of the permeability coefficient is provided by COSMOperm software.
Note,
all described methods can be browsed in the database at https://molmedb.upol.cz/browse/methods. In addition to the ability to browse methods within the classification system, detailed descriptions of each method are also included. This option is illustrated in Figure 3.
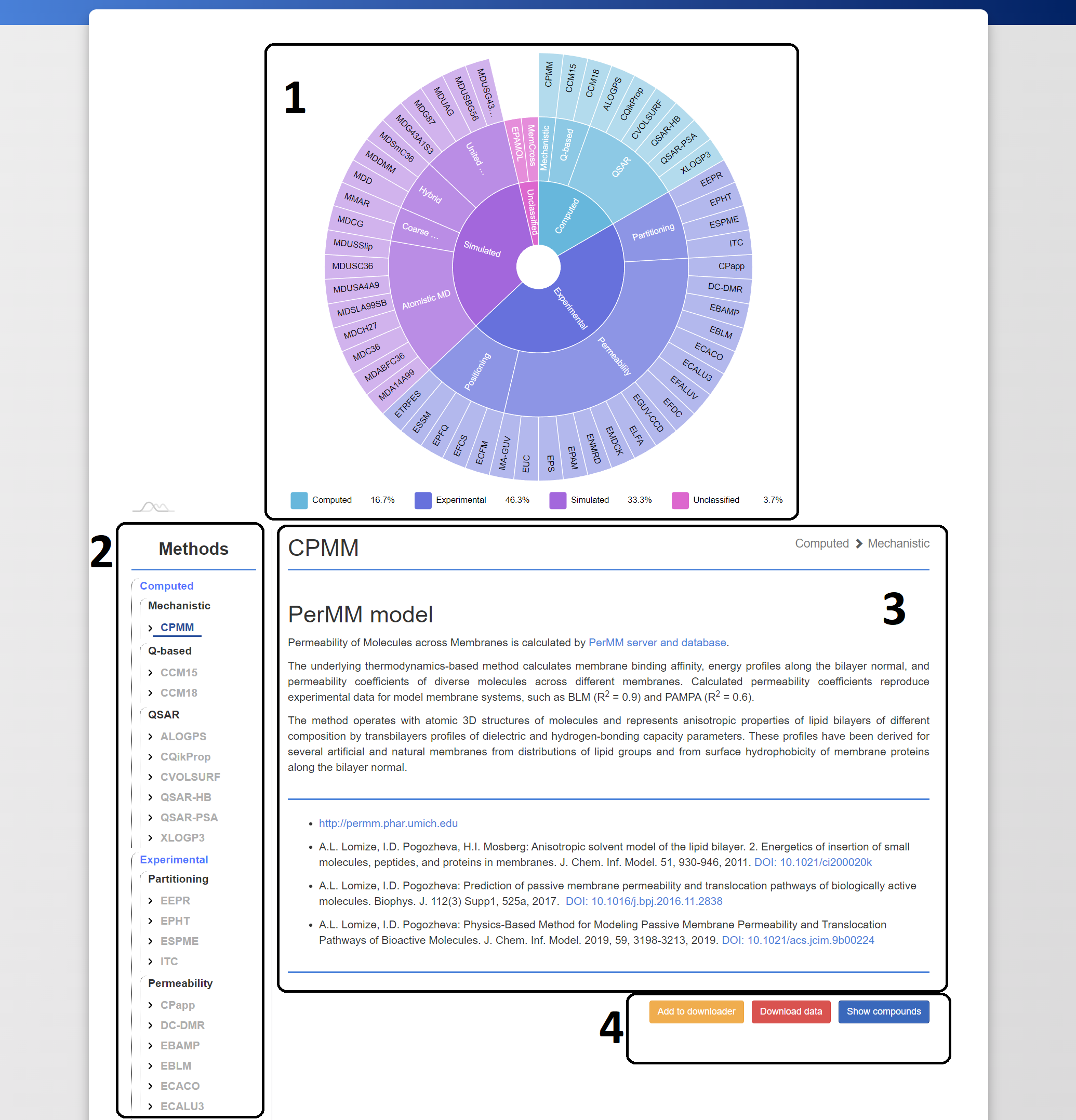
Figure 3. Browsing the methods in MolMeDB. (1) Pie chart of method classification, (2) – list of all available methods, (3) - description of the method, (4) – method-associated actions.
Membranes
Currently (01/2023), there are 40 membranes in MolMeDB. Similarly to the methods, a classification system for membranes was defined with the seven main categories – cell, skin, eye, intestine, brain, sublingual and generic. Each category has common subcategories – experimental and model. Experimental membranes refer to membranes consisting of cells, e.g., membrane SC (stratum corneum) is a part of skin tissue. On the contrary, model membranes are composed of one or more chemical substances and, in general, are much chemically simpler than experimental membranes. An example of a model membrane is CERN. The CERN is a model skin membrane that models the stratum corneum as a bilayer of ceramides. GENER category is used when the literature does not specify the used membrane.
Active interactions
In contrast to passive interactions, active interactions describe a compound's behaviour with membrane transport proteins3. In MolMeDB, there are included:
- Ion channels - potassium, calcium, and sodium ion channels,
- Transporters - solute carriers (SLC), ATP-binding cassette transporters (ABC) and ATPases.
The data about the active interactions was gathered by data mining from PubChem, ChEMBL, Metrabase and IUPHAR in collaboration with A. Tuerková and B. Zdrazil.
Data about the active interaction of the molecule are available in the section Transporters on a compound page. In this section, there is the name of the transporter, UniProt ID, type of interaction, numerical values of interactions and primary and secondary references. Complete list of the defined properties:
- Target – name of the transporter,
- UniProt ID – link to the detailed information about the transporter on the UniProt website,
- Type – word description of active interaction,
- Km – Michaelis constant,
- Ki – inhibition constant,
- IC50 - half maximal inhibitory concentration,
- EC50 - half maximal effective concentration.
Active interactions are described numerically and verbally. The numerical values are the negative decimal logarithm4 of Km, Ki, IC50 and EC50. The type is a one-word description of the interaction used for quick orientation in the section Transporters and acquiring four values – non-substrate, substrate, non-inhibitor, and inhibitor. The value of type is given by the numerical value according to these rules:
- Non-substrate: pKm or pEC50 < 5,
- Substrate: pKm or pEC50 ≥ 5,
- Non-inhibitor: pKi or pIC50 < 5,
- Inhibitor: pKi or pIC50 ≥ 5.
3 Membrane transport proteins are a wide group of various types of proteins which includes porins, channels, transporters, carriers etc. For simplicity, we use only the terms „transporter“ or „channel“.
4 MolMeDB stores the negative decimal logarithms of numerical values in this section, and hence these physical quantities are referred to as pKm, pKi, pIC50 and pEC50.
Note,
transporters can be browsed at https://molmedb.upol.cz/browse/transporters.